Membrane-Protein Modeling
Research in the Delemotte Lab focuses on understanding the molecular basis for the function and modulation of membrane proteins, the molecular machines in the cell membranes that enable cellular communication. To do so, we mainly use computational methods such as molecular dynamics simulations and sequence analysis.
We are big believers in open science and make our simulation and analysis code available on our Lab Github, the data necessary to reproduce our papers on OSF, and post our drafts as soon as they are ready for peer-review on public repositories (BioRXiv or Arxiv).
Visit the project pages below to learn more about the different applications we are interested in!

Principal Investigator
Researchers and PhD Students
Ion Channel Gating
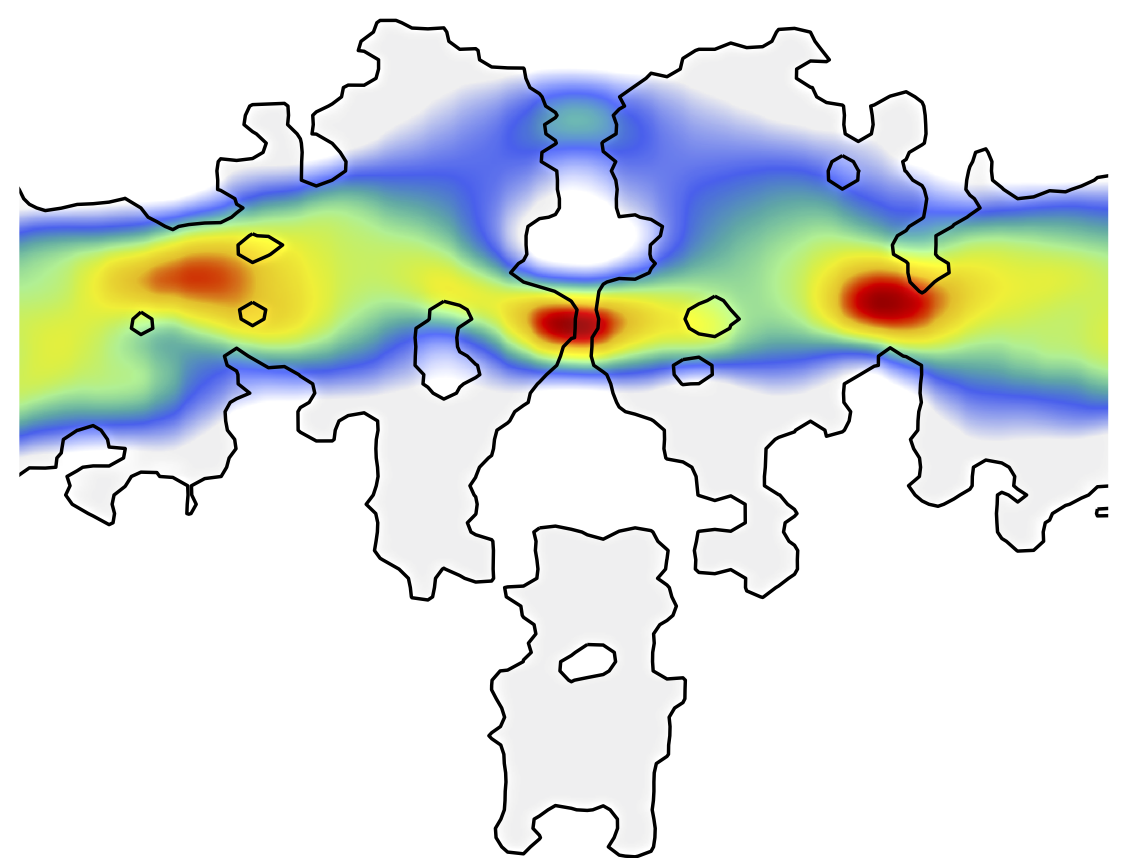
How does membrane potential activate pacemaker channels of the heart? What is the molecular basis of sodium channels inactivation, the process, which prevents prolonged depolarization of membranes?
MD Simulations Analysis

How can one use various methods of data analysis to extract physiologically relevant information from a large ensemble of protein conformations observed in simulations?
Enhanced Sampling
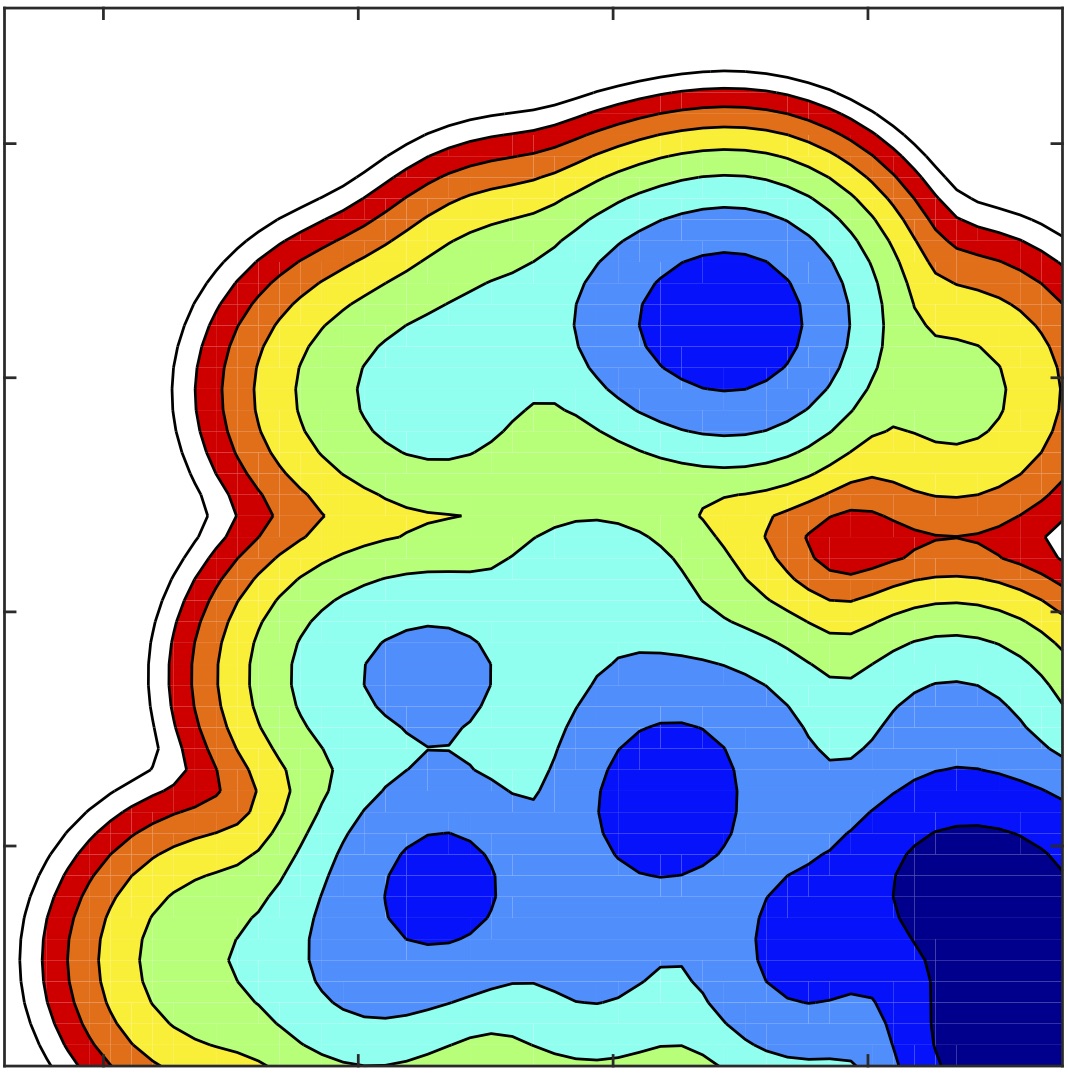
How can one use enhanced sampling techniques to understand biological phenomena, characterize the free energy and the kinetics of biomolecular processes?
GPCRs
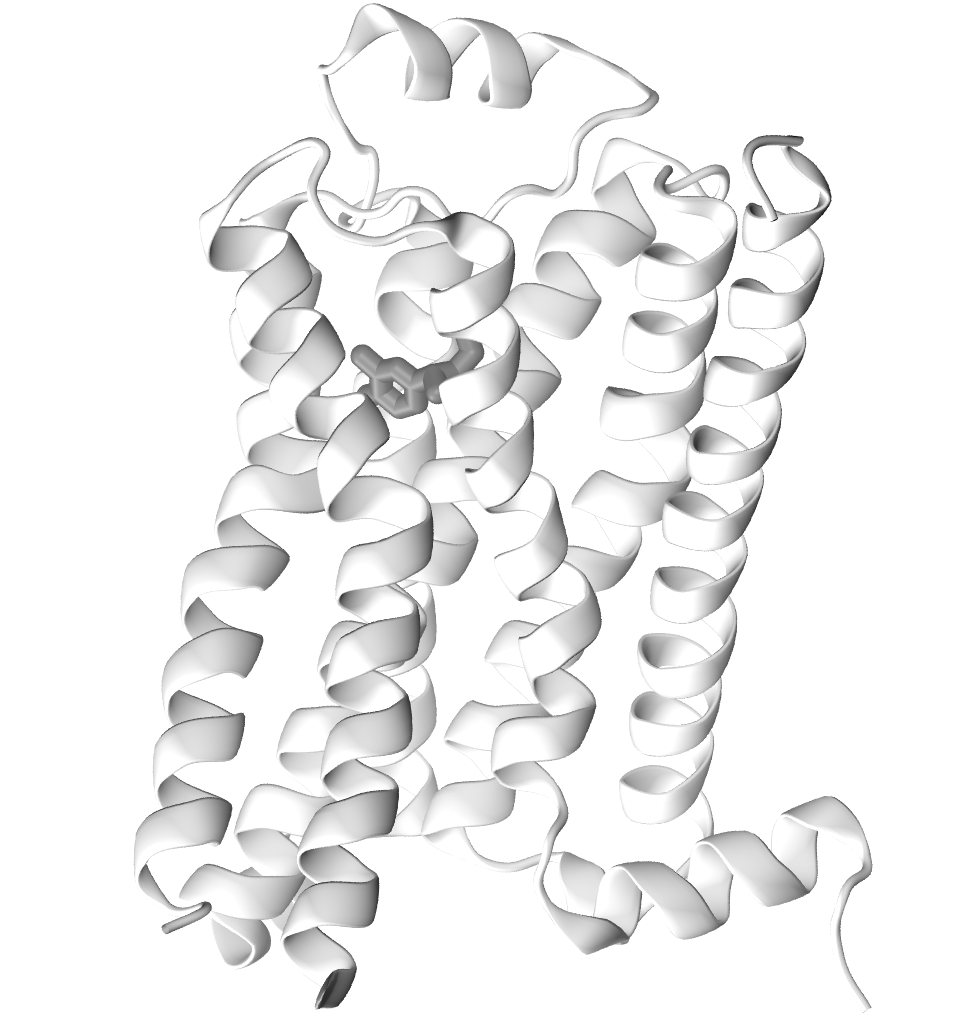
How do GPCRs activate? How is the ligand binding site coupled to the intracellular domain?
Transporter Dynamics
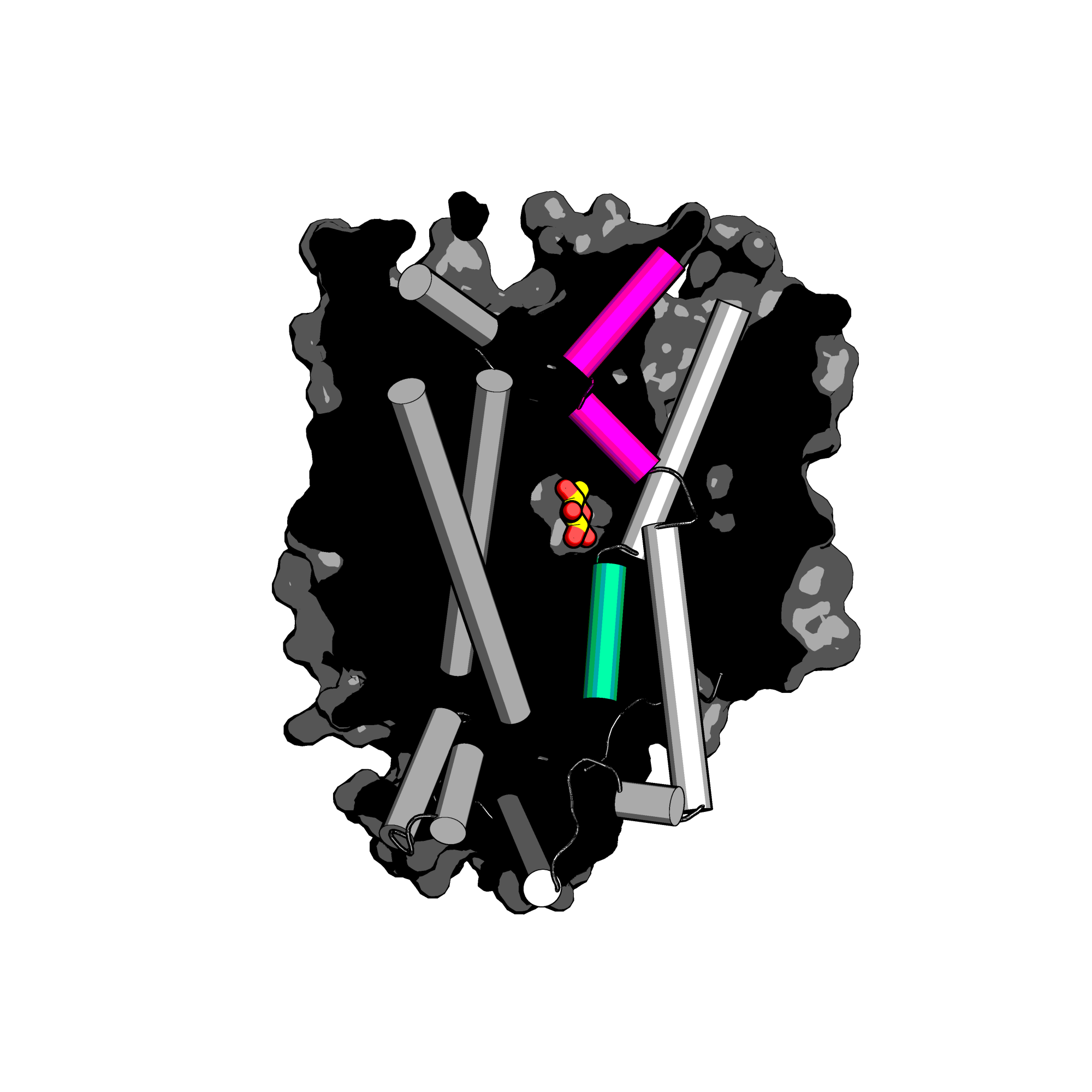
How is substrate recognized in sugar porters? What drives the changes of these proteins through their alternating access mechanism?
Modulation
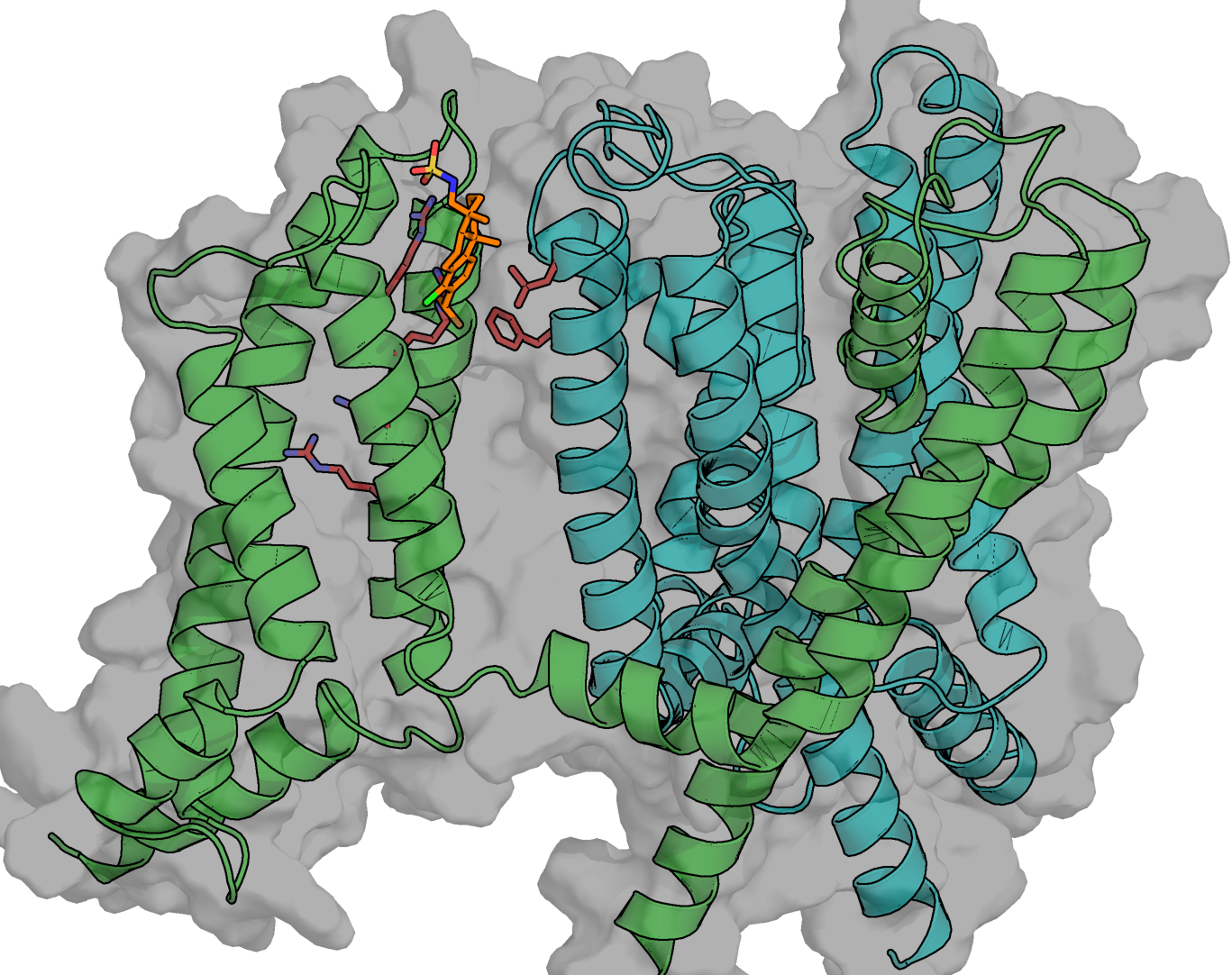
How are membrane proteins modulated by lipids and/or drugs?


You must be logged in to post a comment.