In addition to the positions below, we are always interested in hearing from motivated applicants at join-mbp@scilifelab.se.
Visit the Teams page to learn more about our work and appropriate contacts.
Research Projects in Integrative Structural Biology
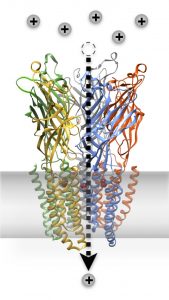
Led by Professor Erik Lindahl and Senior Researcher Reba Howard, the Molecular Biophysics Stockholm Integrative Structural Biology Team combines structural, functional, and computational approaches to characterize complex biomolecules. Members are engaged at various levels in connecting insights between experimental biochemistry data and molecular dynamics simulations. Historically, the team has focused on applications to the rich allostery and pharmaceutical potential of ligand-gated ion channels.
Four types of research projects at the bachelor’s, master’s, doctoral, or postdoctoral levels are currently planned in the following areas:
Cryo-Electron Microscopy of Membrane Proteins
Optimize sample preparation, data collection, and image analysis to elucidate the atomic structure of technically challenging macromolecules, focusing on biochemistry or software methods in cryo-electron microscopy.
Electrophysiology of Novel Receptor Variants
Express and characterize ion channel proteins in heterologous cells, using molecular biology and voltage-clamp recordings to elucidate molecular mechanisms and pharmacology.
Simulating Ion Channel Gating and Modulation
Harness structure-function data to simulate and analyze modeled ion channels, using molecular dynamics to probe structural changes, drug binding, and disruptive mutations.
AI Approaches to Enhance Biomolecular Modeling
Develop and implement machine-learning based analyses to integrate various data types in characterizing structure and dynamics of proteins and other biomolecular complexes.
Applicants should have some theoretical and/or laboratory preparation in biophysics, biochemistry, or related fields, and an enthusiasm for interdisciplinary research and communication.
Formal application periods for PhD students and postdoctoral fellows are anticipated in the coming year. For currently posted positions, please check the Stockholm University or KTH Royal Institute of Technology websites.
Contact join-mbp@scilifelab.se for further details.

You must be logged in to post a comment.